CytoSyn: A State-of-the-Art Diffusion Model for Histopathology Image Generation
As part of its long-term effort to tackle scanner & staining variability, a major hindrance in the deployment of deep-learning-based computational pathology, Owkin is proud to release CytoSyn, a new diffusion model enabling the generation of histopathology images.
CytoSyn is only available to academic and research institutions, for non-commercial use, on HuggingFace: https://huggingface.co/Owkin-Bioptimus/CytoSyn. A publication preprint with more details and experiments will be released soon.
Dataset & Model
CytoSyn is based on the REPA-E architecture [1], and has been trained on around 40M tiles (224x224 images) extracted at 0.5MPP (microns per pixel) from areas containing tissue in ~10k H&E TCGA Diagnostic [2] whole slide images (WSIs). The training tiles have been sampled to follow the distribution of tissue source sites in TCGA WSIs, as to be as diverse as possible. The dataset contains tiles from 32 different indications.
The model consists of 3 different components (totaling 853M parameters):
-> a variational auto-encoder (SD-VAE [3], f8d4, 84M parameters),
-> a transformer-based latent diffusion model (SiT XL/2 [4], 683M parameters),
-> H0-mini [5], fruit of a previous collaboration between Owkin & Bioptimus, used as the conditioning model (ViT-b/14, 86M parameters). H0-mini is a features extractor obtained through the distillation of H-optimus-0 [6], a large state-of-the-art features extractor trained with the self-supervised method from DINOv2 [7]. In addition to guidance, H0-mini was also used as the alignment model in the REPA-E architecture.
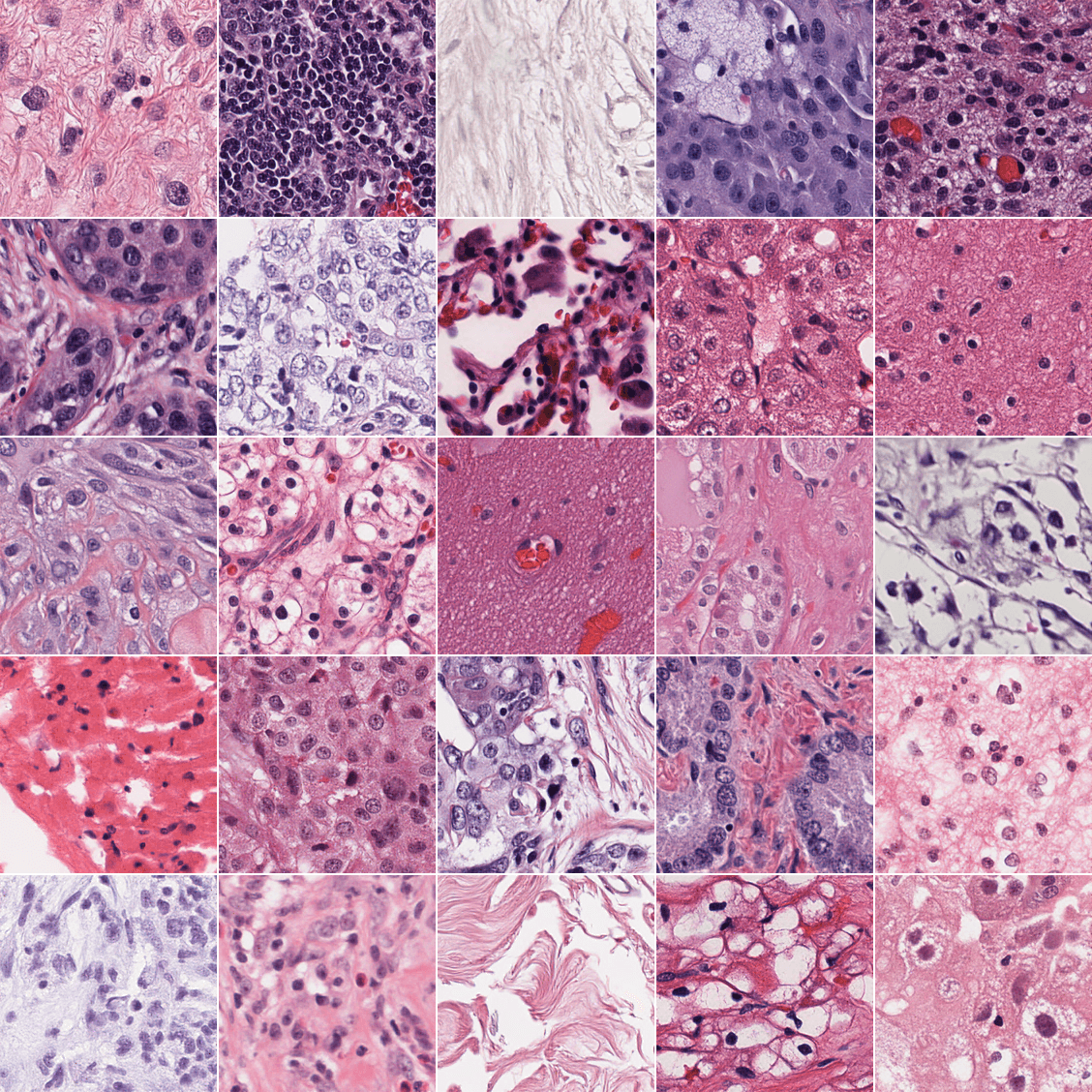
H0-mini guidance enables the generation of tiles conditioned to other tiles' feature vectors:
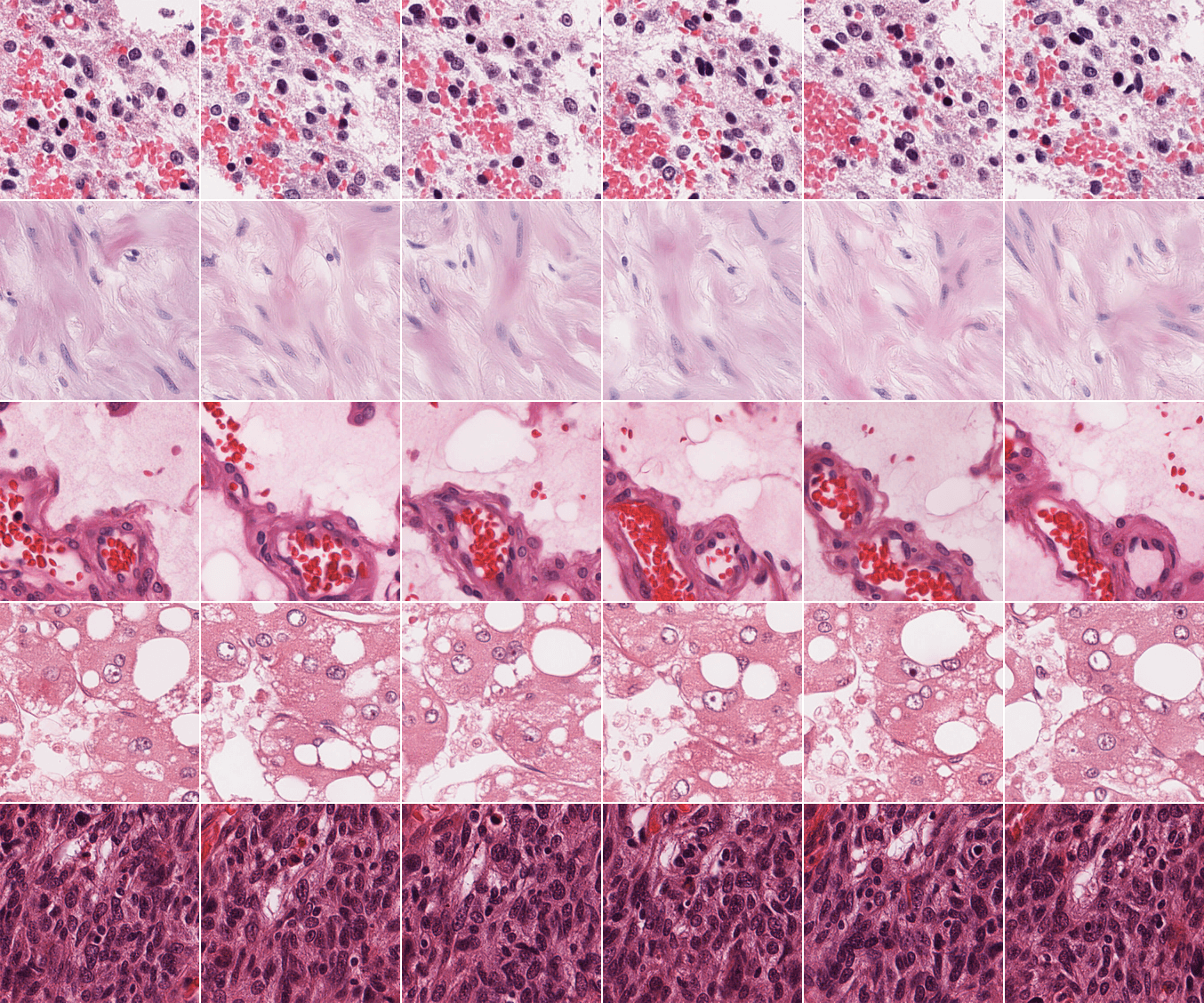
Experiments & Results
We compared our model to PixCell-256 [8], as it is currently the only other publicly released pan-cancer histology diffusion model, to the best of our knowledge. The validation set used consists of 50k tiles coming from TCGA WSIs entirely excluded from the training set. Following Pixcell's methodology, for the conditional FID, we subset 50k tiles from the training set and use their features as conditionings. To align with CytoSyn which uses 250 steps in inference, the images generated with PixCell are also sampled with 250 scheduler's steps (compared to 50 in Pixcell's paper). CytoSyn's results were obtained with the Euler-Maruyama scheduler. Other parameters are kept to their default value in the REPA-E repository, unless specified otherwise.
In addition to the standard Inception-V3 [9] & DINOv2 FID, to provide more fine-grained metrics, we computed FID-scores using several state-of-the-art histopathology-specific features extractors. With the increased realism of the generated images, we posit that histopathology-specific features extractor are going to be able to uncover subtle differences in generated tiles while models trained on ImageNet-like datasets will no longer be able to, as can be seen with the different hyper-parameter sets used to sample conditionally with CytoSyn (last two rows of Table 1.). Outside of this specific scenario, we find a high level of agreement between the different feature extractors.
FID scores assess the overall realism of the generated images, but not the quality of the learned conditional sampling. To measure it, we use the H0-mini features of the validation set to generate a synthetic ‘validation’-like set, and compare the embeddings similarity (for different extractors) between the real and synthetic sets.
Our results show that CytoSyn reaches state-of-the-art results for both FID and cosine similarity.
Note that due to differences in:
-> tiling (the process of extracting tiles from WSIs, includes resizing with potentially different interpolations, extracting in different final formats like jpeg or png),
-> size of generated images (CytoSyn generates 224x224 images while PixCell generates 256x256 images, with resizing operations being known to alter FID scores [10]),
-> training sets (CytoSyn has been trained on TCGA diagnostic data only while PixCell has been trained on a mix of TCGA diagnostic, TCGA fresh frozen, CPTAC [11], GTEx [12] and internal data) and
-> implementation of the performance metrics (PixCell's results were obtained using the Clean-FID implementation [10] while we relied on the implementation from the paper 'Feature Likelihood Divergence: Evaluating the Generalization of Generative Models Using Samples' [13]),
A fair comparison between PixCell and CytoSyn is difficult and it is not possible to rank them conclusively in absolute terms. On the test set of the private PanCan-30M dataset, PixCell reached an Inception-V3 FID of 9.65, in conditional generation. We attribute the gap between PixCell's original results and the results obtained in the conditional setting on our validation set to these discrepancies. In the non-conditional setting, a noticeably higher FID for PixCell was to be expected as it generates images highly realistic but outside of the TCGA data distribution (e.g. CPTAC-like or GTEX-like tiles). These observations highlight the need for a higher level of standardization in the benchmarking process of image generation models applied to histopathology.
Next Steps
Diffusion models can be used to transfer scanner and staining information across tiles while maintaining the biological signal (e.g. position and nature of cells or tissues), with encouraging results as can be seen on the figure below:
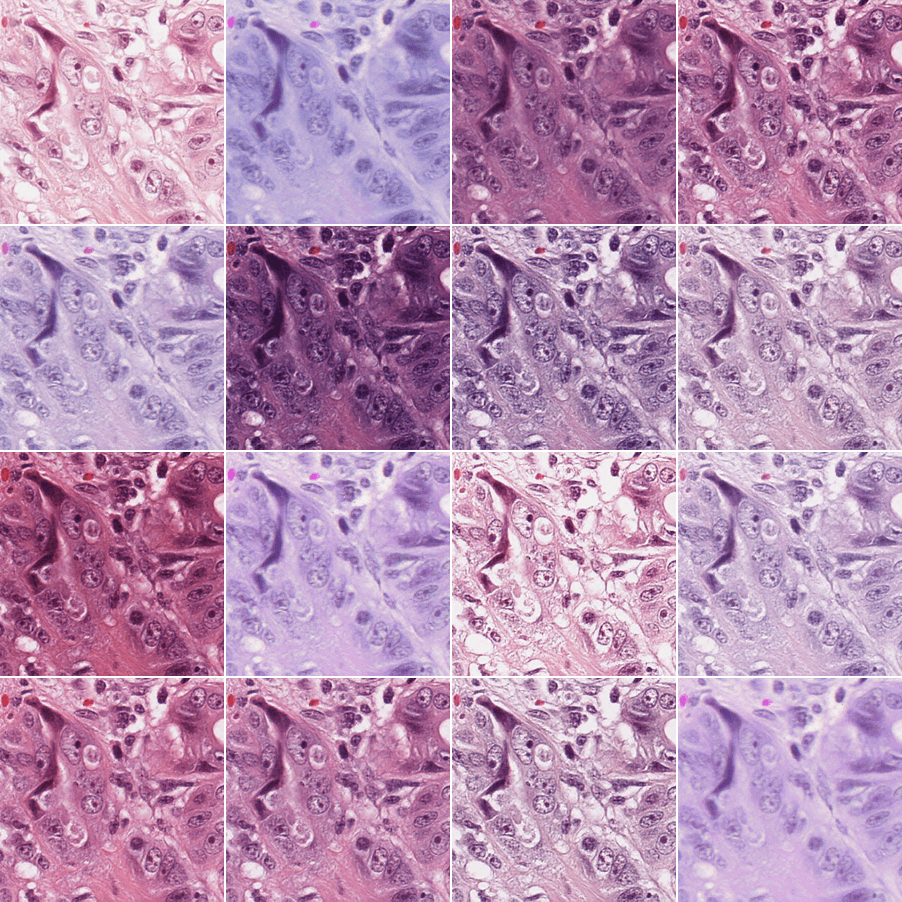
Images generated this way could then be used in a variety of ways to increase robustness to common histopathological domain shifts (e.g. as data augmentation in biomarker prediction models, or as sources for regularization to enforce scanner & staining invariance directly in the training of features extractors, as a stain normalization method, etc.).
References
[1] Leng, X., Singh, J., Hou, Y., Xing, Z., Xie, S., & Zheng, L. (2025). REPA-E: Unlocking VAE for end-to-end tuning with latent diffusion transformers. arXiv preprint arXiv:2504.10483. https://arxiv.org/abs/2504.10483
[2] Weinstein, J. N., Collisson, E. A., Mills, G. B., Shaw, K. R. M., Ozenberger, B. A., Ellrott, K., Shmulevich, I., Sander, C., & Stuart, J. M. (2013). The Cancer Genome Atlas Pan-Cancer analysis project. Nature Genetics, 45(10), 1113–1120. https://doi.org/10.1038/ng.2764
[3] Rombach, R., Blattmann, A., Lorenz, D., Esser, P., & Ommer, B. (2022). High-resolution image synthesis with latent diffusion models. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition (pp. 10684-10695). https://arxiv.org/abs/2112.10752
[4] Ma, N., Goldstein, M., Albergo, M. S., Boffi, N. M., Vanden-Eijnden, E., & Xie, S. (2024, September). Sit: Exploring flow and diffusion-based generative models with scalable interpolant transformers. In European Conference on Computer Vision (pp. 23-40). Cham: Springer Nature Switzerland. https://arxiv.org/abs/2401.08740
[5] Filiot, A., Dop, N., Tchita, O., Riou, A., Dubois, R., Peeters, T., ... & Olivier, A. (2025). Distilling foundation models for robust and efficient models in digital pathology. In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 162-172). Springer. https://arxiv.org/abs/2501.16239
[6] Saillard, C., Jenatton, R., Llinares-López, F., Mariet, Z., Cahané, D., Durand, E., & Vert, J. P. (2024). H-optimus-0 . Bioptimus. https://github.com/bioptimus/releases/tree/main/models/h-optimus/v0
[7] Oquab, M., Darcet, T., Moutakanni, T., Vo, H., Szafraniec, M., Khalidov, V., ... & Bojanowski, P. (2024). DINOv2: Learning Robust Visual Features without Supervision. Transactions on Machine Learning Research Journal. https://arxiv.org/abs/2304.07193
[8] Yellapragada, S., Graikos, A., Li, Z., Triaridis, K., Belagali, V., Kapse, S., ... & Samaras, D. (2025). PixCell: A generative foundation model for digital histopathology images. arXiv preprint arXiv:2506.05127. https://arxiv.org/abs/2506.05127
[9] Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., & Wojna, Z. (2016). Rethinking the Inception Architecture for Computer Vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (pp. 2818-2826). https://arxiv.org/abs/1512.00567
[10] Parmar, G., Zhang, R., & Zhu, J. Y. (2022). On aliased resizing and surprising subtleties in gan evaluation. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition (pp. 11410-11420). IEEE. https://doi.org/10.1109/CVPR52688.2022.01112
[11] Edwards, N. J., Oberti, M., Thangudu, R. R., Cai, S., McGarvey, P. B., Jacob, S., ... & Ketchum, K. A. (2015). The CPTAC data portal: a resource for cancer proteomics research. Journal of proteome research, 14(6), 2707-2713. https://doi.org/10.1021/pr501254j
[12] Lonsdale, J., Thomas, J., Salvatore, M., Phillips, R., Lo, E., Shad, S., ... & Moore, H. F. (2013). The genotype-tissue expression (GTEx) project. Nature genetics, 45(6), 580-585. https://doi.org/10.1038/ng.2653
[13] Jiralerspong, M., Bose, J., Gemp, I., Qin, C., Bachrach, Y., & Gidel, G. (2023). Feature likelihood divergence: evaluating the generalization of generative models using samples. Advances in Neural Information Processing Systems, 36, 33095-33119. https://arxiv.org/abs/2302.04440
[14] Chen, R. J., Ding, T., Lu, M. Y., Williamson, D. F. K., Jaume, G., Song, A. H., ... & Mahmood, F. (2024). Towards a general-purpose foundation model for computational pathology. Nature Medicine, 30(3), 850-862. https://doi.org/10.1038/s41591-024-02857-3
[15] Vorontsov, E., Bozkurt, A., Casson, A., Shaikovski, G., Zelechowski, M., Liu, S., ... & Fuchs, T. J. (2024). Virchow 2: Scaling self-supervised mixed magnification models in pathology. arXiv preprint arXiv:2408.00738. https://arxiv.org/abs/2408.00738
[16] Chen, R. J., Ding, T., Lu, M. Y., Williamson, D. F., Jaume, G., Song, A. H., ... & Mahmood, F. (2024). Towards a general-purpose foundation model for computational pathology. Nature medicine, 30(3), 850-862. https://www.nature.com/articles/s41591-024-02857-3
[17] Filiot, A., Jacob, P., Mac Kain, A., & Saillard, C. (2024). Phikon-v2, a large and public feature extractor for biomarker prediction. arXiv preprint arXiv:2409.09173. https://arxiv.org/abs/2409.09173
[18] Lu, M. Y., Chen, B., Williamson, D. F., Chen, R. J., Liang, I., Ding, T., ... & Mahmood, F. (2024). A visual-language foundation model for computational pathology. Nature medicine, 30(3), 863-874. https://www.nature.com/articles/s41591-024-02856-4




