Evaluating Owkin’s Discovery AI Model: setting a new standard in AI target discovery
New drug targets are needed
Drug discovery continues to face major challenges, particularly in identifying clinically relevant targets. A key issue is the high failure rate of drug candidates in late-stage clinical development, often due to incomplete understanding of disease biology. To address this, Owkin has developed a state-of-the-art Discovery AI model to revolutionize novel target discovery by integrating diverse biomedical datasets.
Owkin’s Discovery AI is powered by MOSAIC, a large-scale initiative led by Owkin that brings together patient samples from world-leading institutions, multi-omics technologies, and spatial profiling. This combination creates one of the most comprehensive multimodal cancer atlases to date.
While the ultimate validation of AI-selected targets must occur in the lab and clinic, this blog presents early performance insights through a model performance study.
How does Owkin’s Target Discovery AI work?
Owkin’s Discovery AI integrates and learns from a wide range of data types, including: spatial transcriptomics, single-cell and bulk RNA-seq, histopathology images and whole-exome sequencing.
Using proprietary and open-source algorithms, we extract biologically meaningful features from these data sources, creating a high-dimensional gene–patient embedding space of over 80 dimensions. These features capture complex biological signals, such as tumor heterogeneity and cell-type–specific gene expression.
These features are then used to predict the true relationships between molecular targets and disease types, as derived from both successful and failed trials: is a given molecular target predicted as a relevant one in a given disease type? This information is sourced from public and commercial databases focused on late-stage clinical trials, and based on this, Owkin’s Discovery AI develops a nuanced understanding of what makes a target likely to progress clinically.
Owkin’s Discovery AI is a pan-cancer model, trained across more than 30 cancer types, and is designed to predict the potential that a target will reach late-phase clinical trials
Read more about how Owkin’s Discovery AI works here
So how does our model perform against well-established baselines?
Comparing proprietary AI models for target identification remains extremely challenging due to differences in data, methodology, lack of transparency, and IP constraints. Additionally, no single public model has been universally adopted as the gold standard for this task.
To compare the performance of Owkin’s Discovery AI, we evaluated its ability to predict whether a molecular target is associated with a drug that has progressed to a given phase in clinical trials. We compared our model against two widely used public resources: the DepMap dataset and the Open Targets platform.
DepMap (Dependency Map) provides essentiality scores, commonly used by the oncology research community to understand which genes cancer cells rely on to survive and grow.. We used these average scores to rank targets from most to least essential in order to find the best potential new therapeutic targets.
Open Targets is widely regarded as a reference platform in the target ID space. It offers gene–disease association scores derived from multiple data sources. To ensure a fair and novelty-focused comparison, we excluded all associations sourced from literature and ChEMBL to avoid potential data leakage from well-studied genes. This is because the availability of molecular evidence is less likely to be influenced by past associations compared to the literature.
For our evaluation, we compared:
- DepMap’s essentiality scores’ ability to rank targets
- Open Targets’ gene–disease association scores’ ability to rank targets
- The first-stage AI predictions from Owkin’s Discovery AI (excluding downstream filters such as safety or competition)
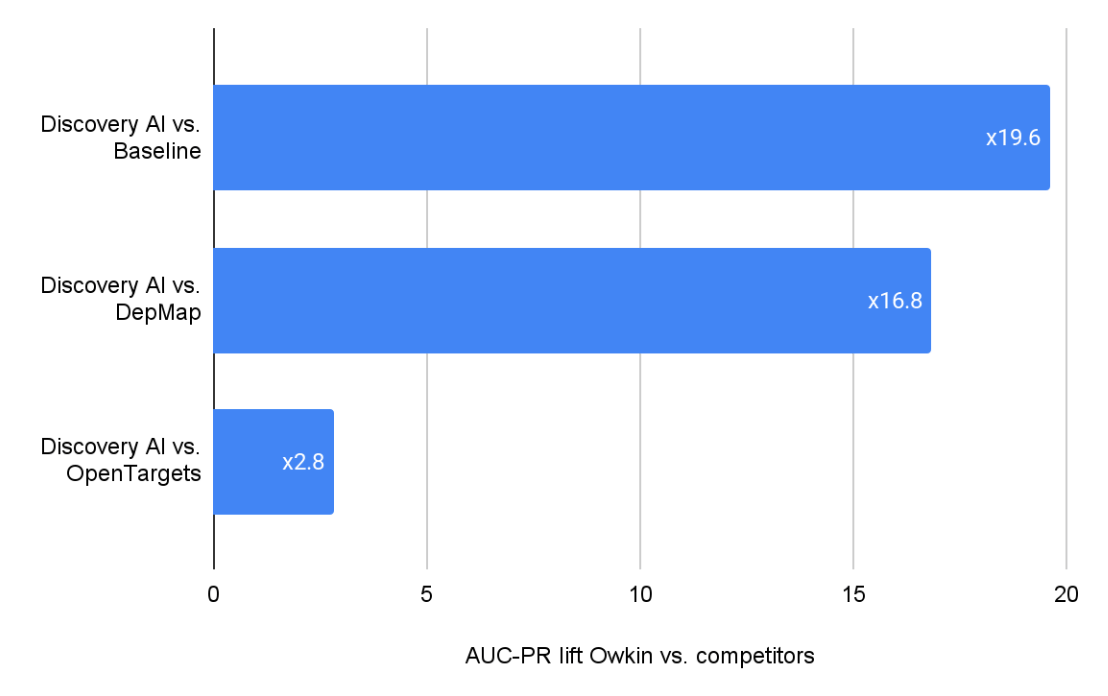
Performance of the three sources’ ability to rank targets was evaluated using the Area Under the Precision-Recall Curve (AUC-PR), a metric well-suited for tasks with class imbalance (i.e., few true positive and/or negative targets among many candidates). We report the lift, defined as the ratio between Owkin’s Discovery AI AUC-PR and that of the baseline models.
Results
Owkin’s Discovery AI consistently outperformed both DepMap and Open Targets in identifying targets across six cancer types from the MOSAIC dataset (Ovarian, Bladder, DLBCL, Mesothelioma, Glioblastoma, and Breast cancer):
- 19.6× more effective than random baseline - which is simply assigning a random score to each gene-indication pair.
- 16.8× more effective than DepMap in retrieving known drug targets in clinical phase 2 studies.
- 2.8× more effective than Open Targets in retrieving known drug targets in clinical phase 2 studies.
These results are averaged across the six indications and show Owkin’s Discovery AI superior ability to identify targets already known to be associated with clinical-phase II drugs.
The metrics presented reflect the average performance across these six indications.
Notably, Owkin’s Discovery AI’s strong performance is significantly enhanced by features from spatial transcriptomics, particularly those describing the spatial relationship of targets to Tertiary Lymphoid Structures (TLS) in the tumor microenvironment.
Impact - A new era of target discovery
Owkin’s Discovery AI demonstrates a clear step forward in AI-driven drug discovery. By integrating multimodal biological data and learning from clinical outcomes across diverse indications, based on this model comparison it can:
- Prioritize novel targets overlooked by traditional tools, specially in the tumour microenvironment
- Reduce attrition in the drug development pipeline, by uncovering more clinically relevant targets
As AI continues to transform biomedicine, Owkin’s Discovery AI provides a compelling case for how well-trained models, when evaluated transparently, can unlock more effective and personalized cancer therapies.
At Owkin, we are packaging high-quality multimodal data together with key AI tools that power our Discovery AI, alongside others developed over the past decade, into our co-pilot for biomedical research: Owkin K-Pro. This platform enables researchers to analyze multimodal datasets and extract the same AI-derived features described above, generating meaningful insights from day one. By helping identify the right populations and relevant biomarkers, Owkin K-Pro accelerates drug discovery and development and supports the design of more targeted and effective clinical trials.




